How do I install biopython in anaconda?
10,216
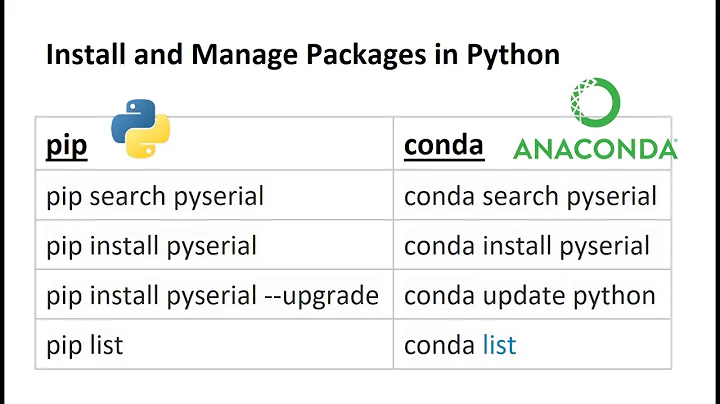
Solution 1
Package maintainers recommend using (in the terminal):
conda install -c conda-forge biopython
We deliberately recommend using Biopython from the conda-forge channel, as this is usually up to date and covers Windows, Mac OS X and Linux. The default Conda channel does have Biopython, but is often out of date. https://biopython.org/wiki/Packages
Solution 2
Open the terminal and export your path to anaconda:
export PATH=~/anaconda3/bin:$PATH
Then type:
conda install -c anaconda biopython
Solution 3
Install Anaconda Navigator, go to "Environments" and select the appropriate environment (base or your own) and click "not installed". Scroll down to biopython click the box and then install...
Related videos on Youtube
Author by
Jaswant S
Updated on June 04, 2022Comments
-
 Jaswant S almost 2 years
Jaswant S almost 2 yearsI get SyntaxError: invalid syntax
while trying to install biopython using the following command.
conda install -c anaconda biopythonCould you please help me install biopython in anaconda (3) ?
-
 FHTMitchell over 5 yearsYou're supposed to put that command into the terminal, not the python interpreter.
FHTMitchell over 5 yearsYou're supposed to put that command into the terminal, not the python interpreter. -
E.Serra over 5 yearsHahahahaha sorry this is the funniest comment ever, +1, and yes he is right you install it through command line
-
 Jaswant S over 5 yearsBut after installation, I get ModuleNotFoundError: No module named 'biopython
Jaswant S over 5 yearsBut after installation, I get ModuleNotFoundError: No module named 'biopython -
 Maximilian Peters over 5 yearsdid you try importing it via
Maximilian Peters over 5 yearsdid you try importing it viaimport Bio?
-