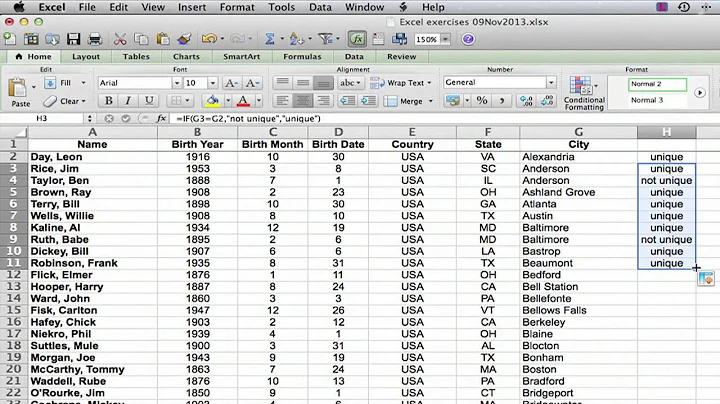
Count number of occurences for each unique value
Solution 1
Perhaps table is what you are after?
dummyData = rep(c(1,2, 2, 2), 25)
table(dummyData)
# dummyData
# 1 2
# 25 75
## or another presentation of the same data
as.data.frame(table(dummyData))
# dummyData Freq
# 1 1 25
# 2 2 75
Solution 2
If you have multiple factors (= a multi-dimensional data frame), you can use the dplyr package to count unique values in each combination of factors:
library("dplyr")
data %>% group_by(factor1, factor2) %>% summarize(count=n())
It uses the pipe operator %>% to chain method calls on the data frame data.
Solution 3
It is a one-line approach by using aggregate.
> aggregate(data.frame(count = v), list(value = v), length)
value count
1 1 25
2 2 75
Solution 4
table() function is a good way to go, as Chase suggested. If you are analyzing a large dataset, an alternative way is to use .N function in datatable package.
Make sure you installed the data table package by
install.packages("data.table")
Code:
# Import the data.table package
library(data.table)
# Generate a data table object, which draws a number 10^7 times
# from 1 to 10 with replacement
DT<-data.table(x=sample(1:10,1E7,TRUE))
# Count Frequency of each factor level
DT[,.N,by=x]
Solution 5
length(unique(df$col)) is the most simple way I can see.
Related videos on Youtube
gakera
Updated on September 23, 2020Comments
-
gakera almost 4 years
Let's say I have:
v = rep(c(1,2, 2, 2), 25)Now, I want to count the number of times each unique value appears.
unique(v)returns what the unique values are, but not how many they are.> unique(v) [1] 1 2I want something that gives me
length(v[v==1]) [1] 25 length(v[v==2]) [1] 75but as a more general one-liner :) Something close (but not quite) like this:
#<doesn't work right> length(v[v==unique(v)]) -
gakera over 13 yearsAh, yes, I can use this, with some slight modification: t(as.data.frame(table(v))[,2]) is exactly what I need, thank you
-
Brian Diggs about 11 yearsor
ddply(data_frame, .(v), count). Also worth making it explicit that you need alibrary("plyr")call to makeddplywork. -
Museful almost 11 yearsI used to do this awkwardly with
hist.tableseems quite a bit slower thanhist. I wonder why. Can anyone confirm? -
Torvon over 9 yearsChase, any chance to order by frequency? I have the exact same problem, but my table has roughly 20000 entries and I'd like to know how frequent the most common entries are.
-
Chase over 9 years@Torvon - sure, just use
order()on the results. i.e.x <- as.data.frame(table(dummyData)); x[order(x$Freq, decreasing = TRUE), ] -
 Gregor Thomas almost 9 yearsSeems strange to use
Gregor Thomas almost 9 yearsSeems strange to usetransforminstead ofmutatewhen usingplyr. -
 Deep North over 6 yearsThis method is not good, it is only fit for very few data with a lot of repeated, it will not fit a lot of continous data with few duplicated records.
Deep North over 6 yearsThis method is not good, it is only fit for very few data with a lot of repeated, it will not fit a lot of continous data with few duplicated records. -
 Peter over 5 yearsTo count the number of levels you may also use
Peter over 5 yearsTo count the number of levels you may also uselapply(DF, function(x) length(table(x))) -
gakera almost 4 yearsR has probably evolved a lot in the last 10 years, since I asked this question.
-
David almost 4 yearsAlternatively, and a bit shorter:
data %>% count(factor1, factor2) -
 Martin over 3 yearsOne-liner indeed instead of using unique() + something else. Wonderful!
Martin over 3 yearsOne-liner indeed instead of using unique() + something else. Wonderful! -
dsg38 over 2 yearsNB: This doesn't include the NA values
-
vonjd about 2 yearsaggregate is underappreciated!