Error code 100 fitting exp distribution using fitdist in r
Assuming this is fitdist from fitdistrplus package, I can duplicate your error:
> fitdist(x41, "exp", method="mle")
<simpleError in optim(par = vstart, fn = fnobj, fix.arg = fix.arg, obs = data, gr = gradient, ddistnam = ddistname, hessian = TRUE, method = meth, lower = lower, upper = upper, ...): non-finite finite-difference value [1]>
Error in fitdist(x41, "exp", method = "mle") :
the function mle failed to estimate the parameters,
with the error code 100
but there's some large numbers in your data... maybe if we scale it all down by a factor...
> fitdist(x41/10000, "exp", method="mle")
Fitting of the distribution ' exp ' by maximum likelihood
Parameters:
estimate Std. Error
rate 7.1417 1.683315
Well that seemed to work. Let's scale by a bit less:
> fitdist(x41/1000, "exp", method="mle")
Fitting of the distribution ' exp ' by maximum likelihood
Parameters:
estimate Std. Error
rate 0.71417 0.1683312
Right. Divide by a thousand works. Let's keep going:
> fitdist(x41/100, "exp", method="mle")
Fitting of the distribution ' exp ' by maximum likelihood
Parameters:
estimate Std. Error
rate 0.071417 0.01682985
Fine.
> fitdist(x41/10, "exp", method="mle")
Fitting of the distribution ' exp ' by maximum likelihood
Parameters:
estimate Std. Error
rate 0.0071417 0.001649523
So scaling the data by 1/10 works, and you can see how the estimate and SE scale. Let's go one more step:
> fitdist(x41/1, "exp", method="mle")
<simpleError in optim(par = vstart, fn = fnobj, fix.arg = fix.arg, obs = data, gr = gradient, ddistnam = ddistname, hessian = TRUE, method = meth, lower = lower, upper = upper, ...): non-finite finite-difference value [1]>
Error in fitdist(x41/1, "exp", method = "mle") :
the function mle failed to estimate the parameters,
with the error code 100
Crunch. It looks like some numerical stability problem with the underlying algorithm. If its taking exponentials of your data at any point then maybe it hits something indistinguishable from infinity. Like:
> exp(x41)
[1] Inf 2.100274e+132 Inf Inf Inf
[6] Inf 3.757545e+152 5.096228e+47 4.064401e+199 5.776191e+05
[11] 1.033895e+00 Inf Inf Inf 9.145540e+40
[16] 3.323969e+06 1.195135e+118 2.638092e+205
But scale by ten and the maths can cope, just about (E+256!!!)
> exp(x41/10)
[1] 2.552833e+121 1.706977e+13 1.032728e+121 1.367817e+256 1.907002e+190
[6] 1.459597e+51 1.809216e+15 5.898273e+04 9.139021e+19 3.768462e+00
[11] 1.003339e+00 5.727429e+36 4.184491e+160 2.094645e+66 1.247731e+04
[16] 4.489166e+00 6.423056e+11 3.484408e+20
Related videos on Youtube
Judy S
Updated on July 25, 2022Comments
-
 Judy S almost 2 years
Judy S almost 2 yearsI am trying to fit an exponential distribution to my data but I get the error below
"Error in fitdist(x41, "exp", method = "mle") : the function mle failed to estimate the parameters, with the error code 100"I have tried mme which works but I have other distributions with mle so I need mle in exponential distribution as well. I have been stuck for days. Can anyone help me, please?
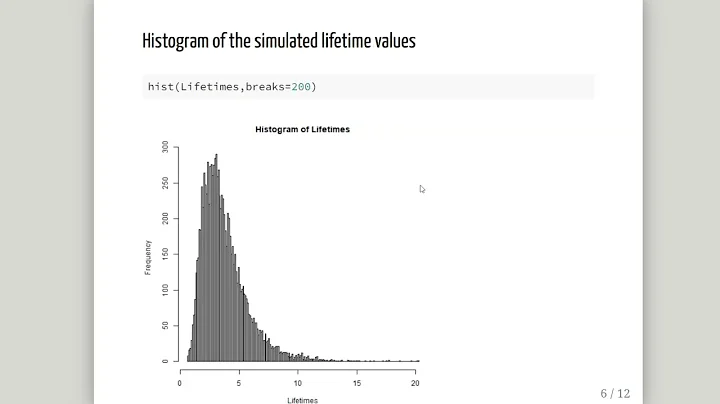
My data looks like this.
2795.5 304.6833 2786.45 5897.75 4381.367 1178.1 351.3167 109.85 459.6167 13.26667 0.033333 846.3833 3698.45 1527.1 94.31667 15.01667 271.8833 473This is my code
ExpMle41 <- fitdist(x41, "exp", method="mle") ExpMle41 plot(ExpMle41)Any help will be greatly appreciated. Thank you.
-
 Judy S over 5 yearsThanks a lot for the help. I really appreciate it.
Judy S over 5 yearsThanks a lot for the help. I really appreciate it.