How do I standardize a matrix?
Solution 1
The following subtracts the mean of A from each element (the new mean is 0), then normalizes the result by the standard deviation.
import numpy as np
A = (A - np.mean(A)) / np.std(A)
The above is for standardizing the entire matrix as a whole, If A has many dimensions and you want to standardize each column individually, specify the axis:
import numpy as np
A = (A - np.mean(A, axis=0)) / np.std(A, axis=0)
Always verify by hand what these one-liners are doing before integrating them into your code. A simple change in orientation or dimension can drastically change (silently) what operations numpy performs on them.
Solution 2
import scipy.stats as ss
A = np.array(ss.zscore(A))
Solution 3
from sklearn.preprocessing import StandardScaler
standardized_data = StandardScaler().fit_transform(your_data)
Example:
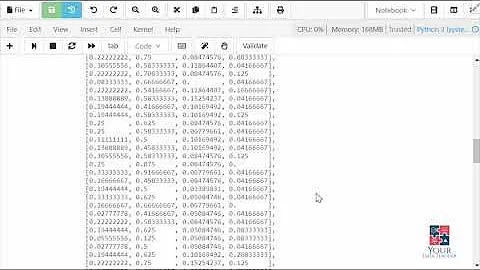
>>> import numpy as np
>>> from sklearn.preprocessing import StandardScaler
>>> data = np.random.randint(25, size=(4, 4))
>>> data
array([[17, 12, 4, 17],
[ 1, 16, 19, 1],
[ 7, 8, 10, 4],
[22, 4, 2, 8]])
>>> standardized_data = StandardScaler().fit_transform(data)
>>> standardized_data
array([[ 0.63812398, 0.4472136 , -0.718646 , 1.57786412],
[-1.30663482, 1.34164079, 1.55076242, -1.07959124],
[-0.57735027, -0.4472136 , 0.18911737, -0.58131836],
[ 1.24586111, -1.34164079, -1.02123379, 0.08304548]])
Works well on large datasets.
Solution 4
Use sklearn.preprocessing.scale.
http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.scale.html
Here is an example.
>>> from sklearn import preprocessing
>>> import numpy as np
>>> X_train = np.array([[ 1., -1., 2.],
... [ 2., 0., 0.],
... [ 0., 1., -1.]])
>>> X_scaled = preprocessing.scale(X_train)
>>> X_scaled
array([[ 0. ..., -1.22..., 1.33...],
[ 1.22..., 0. ..., -0.26...],
[-1.22..., 1.22..., -1.06...]])
Related videos on Youtube
pnodbnda
Updated on July 09, 2022Comments
-
pnodbnda almost 2 years
Basically, take a matrix and change it so that its mean is equal to 0 and variance is 1. I'm using numpy's arrays so if it can already do it it's better, but I can implement it myself as long as I can find an algorithm.
edit: nvm nimrodm has a better implementation
-
Karl Knechtel over 13 yearsDefine "change". What if, say, we just replace the matrix with the identity matrix or something? What kinds of transformations are OK?
-
 Drew Hall over 13 yearsJust out of curiosity, why do you need to do this?
Drew Hall over 13 yearsJust out of curiosity, why do you need to do this? -
pnodbnda over 13 yearsI'm trying to implement a computer vision algorithm that asks for this operation to be performed in the intermediate steps. I think it's because it's a requirement for PCA but I'm not sure.
-
 John Alexiou over 6 yearsWould you consider accepting the answer by @nimrodm so I can delete mine?
John Alexiou over 6 yearsWould you consider accepting the answer by @nimrodm so I can delete mine?
-
-
 Jean-François Fabre over 7 yearsyou could use ctrl+k to indent everything instead of backticks.
Jean-François Fabre over 7 yearsyou could use ctrl+k to indent everything instead of backticks. -
Ciprian Tomoiagă over 7 yearsyou may want to update
Aonly wherestd(A) > 0to avoid division by zero andNaNvalues -
Nematode7 about 7 yearsIs this possible where A is represented as a list of lists?
-
 kingledion about 7 years@Neamah Why not just convert to a numpy array?
kingledion about 7 years@Neamah Why not just convert to a numpy array? -
user3585984 about 5 yearsAdding to @nimrodm's answer, this can be implemented in numpy as follows import numpy as np meanArr = np.mean(A) standardized_arr = (A-meanArr)/np.std(A)