How to join files with required columns in linux?
GNU awk is used. I put this command in the bash script. It will be more convenient.
Usage: ./join_files.sh or, for pretty printing, do: ./join_files.sh | column -t.
#!/bin/bash
gawk '
NR == 1 {
PROCINFO["sorted_in"] = "@ind_num_asc";
header = $1;
}
FNR == 1 {
file = gensub(/.*\/([^.]*)\..*/, "\\1", "g", FILENAME);
header = header OFS file;
}
FNR > 1 {
arr[$1] = arr[$1] OFS $5;
}
END {
print header;
for(i in arr) {
print i arr[i];
}
}' results/*.genes.results
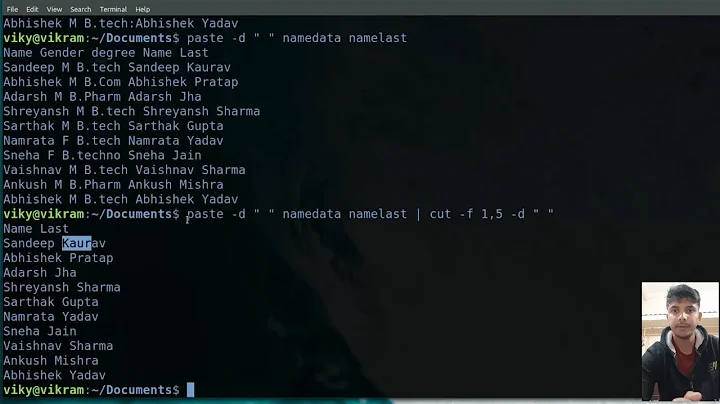
Output (I created three files with the same content for testing)
$ ./join_files.sh | column -t
gene_id TB1 TB2 TB3
ENSG00000000003 1.00 1.00 1.00
ENSG00000000005 0.00 0.00 0.00
ENSG00000000419 1865.00 1865.00 1865.00
ENSG00000000457 1521.00 1521.00 1521.00
ENSG00000000460 1860.00 1860.00 1860.00
ENSG00000000938 6846.00 6846.00 6846.00
ENSG00000000971 0.00 0.00 0.00
ENSG00000001036 1358.00 1358.00 1358.00
ENSG00000001084 1178.00 1178.00 1178.00
Explanation - the same code with comments added. Also, look at the man gawk.
gawk '
# NR - the total number of input records seen so far.
# If the total line number is equal 1
NR == 1 {
# If the "sorted_in" element exists in PROCINFO, then its value controls
# the order in which array elements are traversed in the (for in) loop.
# else the order is undefined.
PROCINFO["sorted_in"] = "@ind_num_asc";
# Each field in the input record may be referenced by its position: $1, $2, and so on.
# $1 - is the first field or the first column.
# The first field in the first line is the "gene_id" word;
# Assign it to the header variable.
header = $1;
}
# FNR - the input record number in the current input file.
# NR is the total lines counter, FNR is the current file lines counter.
# FNR == 1 - if it is the first line of the current file.
FNR == 1 {
# remove from the filename all unneeded parts by the "gensub" function
# was - results/TB1.genes.results
# become - TB1
file = gensub(/.*\/([^.]*)\..*/, "\\1", "g", FILENAME);
# and add it to the header variable, concatenating it with the
# previous content of the header, using OFS as delimiter.
# OFS - the output field separator, a space by default.
header = header OFS file;
}
# some trick is used here.
# $1 - the first column value - "gene_id"
# $5 - the fifth column value - "expected_count"
FNR > 1 {
# create array with "gene_id" indexes: arr["ENSG00000000003"], arr["ENSG00000000419"], so on.
# and add "expected_count" values to it, separated by OFS.
# each time, when the $1 equals to the specific "gene_id", the $5 value will be
# added into this array item.
# Example:
# arr["ENSG00000000003"] = 1.00
# arr["ENSG00000000003"] = 1.00 2.00
# arr["ENSG00000000003"] = 1.00 2.00 3.00
arr[$1] = arr[$1] OFS $5;
}
END {
print header;
for(i in arr) {
print i arr[i];
}
}' results/*.genes.results
Related videos on Youtube
stack_learner
Updated on September 18, 2022Comments
-
stack_learner over 1 year
I have many files like following in a directory "results"
58052 results/TB1.genes.results 198003 results/TB1.isoforms.results 58052 results/TB2.genes.results 198003 results/TB2.isoforms.results 58052 results/TB3.genes.results 198003 results/TB3.isoforms.results 58052 results/TB4.genes.results 198003 results/TB4.isoforms.resultsFor eg: TB1.genes.results file looks like following:
gene_id transcript_id(s) length effective_length expected_count TPM FPKM ENSG00000000003 ENST00000373020,ENST00000494424,ENST00000496771,ENST00000612152,ENST00000614008 2206.00 1997.20 1.00 0.00 0.01 ENSG00000000005 ENST00000373031,ENST00000485971 940.50 731.73 0.00 0.00 0.00 ENSG00000000419 ENST00000371582,ENST00000371584,ENST00000371588,ENST00000413082,ENST00000466152,ENST00000494752 977.15 768.35 1865.00 14.27 37.82 ENSG00000000457 ENST00000367770,ENST00000367771,ENST00000367772,ENST00000423670,ENST00000470238 3779.11 3570.31 1521.00 2.50 6.64 ENSG00000000460 ENST00000286031,ENST00000359326,ENST00000413811,ENST00000459772,ENST00000466580,ENST00000472795,ENST00000481744,ENST00000496973,ENST00000498289 1936.74 1727.94 1860.00 6.33 16.77 ENSG00000000938 ENST00000374003,ENST00000374004,ENST00000374005,ENST00000399173,ENST00000457296,ENST00000468038,ENST00000475472 2020.10 1811.30 6846.00 22.22 58.90 ENSG00000000971 ENST00000359637,ENST00000367429,ENST00000466229,ENST00000470918,ENST00000496761,ENST00000630130 2587.83 2379.04 0.00 0.00 0.00 ENSG00000001036 ENST00000002165,ENST00000367585,ENST00000451668 1912.64 1703.85 1358.00 4.69 12.42 ENSG00000001084 ENST00000229416,ENST00000504353,ENST00000504525,ENST00000505197,ENST00000505294,ENST00000509541,ENST00000510837,ENST00000513939,ENST00000514004,ENST00000514373,ENST00000514933,ENST00000515580,ENST00000616923 2333.50 2124.73 1178.00 3.26 8.64Other files also has the same columns. To join all "genes.results" with "gene_id" and "expected_count" columns into one text file I gave the following command.
paste results/*.genes.results | tail -n+2 | cut -f1,5,12,19,26 > final.genes.rsem.txt [-f1 (gene_id), 5 (expected_count column from TB1.genes.results), 12 (expected_count column from TB2.genes.results), 19 (expected_count column from TB3.genes.results), 26 (expected_count column from TB4.genes.results)]"final.genes.rsem.txt" has, selected gene_id and expected_count columns from every file.
ENSG00000000003 1.00 0.00 3.00 2.00 ENSG00000000005 0.00 0.00 0.00 0.00 ENSG00000000419 1865.00 1951.00 5909.00 8163.00 ENSG00000000457 1521.00 1488.00 849.00 1400.00 ENSG00000000460 1860.00 1616.00 2577.00 2715.00 ENSG00000000938 6846.00 5298.00 1.00 2.00 ENSG00000000971 0.00 0.00 6159.00 7069.00 ENSG00000001036 1358.00 1186.00 6196.00 7009.00 ENSG00000001084 1178.00 1186.00 631.00 1293.00My question is - As I have only few samples I gave the column number in the command [like this in "cut" -f1,5,12,19,26]. What I should do if I have more than 100 samples. How can I join them with required columns?
-
 RomanPerekhrest over 6 yearsthe input file contents is unclear. some values delimited with commas, some - with spaces. Besides, what are the values of
RomanPerekhrest over 6 yearsthe input file contents is unclear. some values delimited with commas, some - with spaces. Besides, what are the values oflengthcolumn? -
MiniMax over 6 years@RomanPerekhrest I understood this file format (not right away) - the delimiter is the space only. So, it have 7 fields. The comma is not delimiter, but the part of the field content. Thus, gene_id =
ENSG00000000003, transcript_id(s) =ENST00000373020,ENST00000494424,ENST00000496771,ENST00000612152,ENST00000614008, length =2206.00, so on.
-
-
stack_learner over 6 yearsIs everything in one line?
-
MiniMax over 6 years@user3351523 Yes, I just split command to multiple lines for readability. Check, does it work.
-
MiniMax over 6 years@user3351523 I simplified the command.
-
stack_learner over 6 yearsNo it doesn't work. It doesn't give me the output I want.
-
MiniMax over 6 years@user3351523 I understood incorrectly your question. Now, I am grasping it.
-
John Smith over 6 yearsTry this
awk '{print $1, $5}' results/*.genes.results. It should print you only two columns, gene_id and expected_count. Is this what you want? -
stack_learner over 6 yearsI need gene_id as 1st column, expected_count column from TB1.genes.results as 2nd column, expected_count column from TB2.genes.results as 3rd column, expected_count column from TB3.genes.results as 4th column, expected_count column from TB4.genes.results as 5th column. I did this by using the command I mentioned in the above question. But would like to know how I can do this with 100 files.
-
MiniMax over 6 years@user3351523 Another try :) Check it now.
-
John Smith over 6 yearsdo you have the same gene_id's in corresponding lines of TB1.gene.results, TB2.gene.results, TB3.gene.results and so on? for example, if you have gene_id ENSG00000000003 in the first line of TB1, does that mean that in TB2 and TB3 you also have first line with the same gene_id ENSG00000000003?
-
stack_learner over 6 yearsYes, gene_id column is same in all the files.
-
stack_learner over 6 yearsDo i need to give this in a script or just on command line? How to save the output in a file?
-
MiniMax over 6 years@user3351523 As you want. It can be run as command in the terminal directly (just copy this command and paste it to the command line, then press Enter) or it can be putted in the bash script. I added saving output to the file. Also, some corrections added - headers skipping in the each file.
-
stack_learner over 6 yearsYes it worked, but I have a small question. Is it possible to give the column names for that? Like - gene_id, TB1, TB2, TB3, TB4
-
stack_learner over 6 yearscould you please explain the code once. What is FNR and OFS?
-
MiniMax over 6 years@user3351523 I can, but few hours later. I will add the explanation and try to implement your request about filenames in the header.
-
stack_learner over 6 yearsThank you very much for the script and it worked perfectly and thanks a lot for explanation.