How to normalize a 2-dimensional numpy array in python less verbose?
Solution 1
Broadcasting is really good for this:
row_sums = a.sum(axis=1)
new_matrix = a / row_sums[:, numpy.newaxis]
row_sums[:, numpy.newaxis] reshapes row_sums from being (3,) to being (3, 1). When you do a / b, a and b are broadcast against each other.
You can learn more about broadcasting here or even better here.
Solution 2
Scikit-learn offers a function normalize() that lets you apply various normalizations. The "make it sum to 1" is called L1-norm. Therefore:
from sklearn.preprocessing import normalize
matrix = numpy.arange(0,27,3).reshape(3,3).astype(numpy.float64)
# array([[ 0., 3., 6.],
# [ 9., 12., 15.],
# [ 18., 21., 24.]])
normed_matrix = normalize(matrix, axis=1, norm='l1')
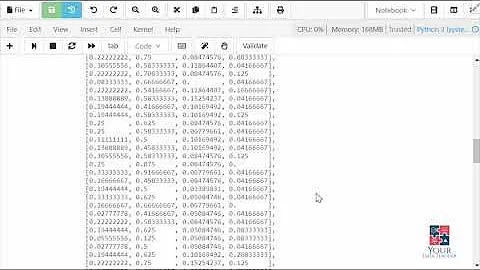
# [[ 0. 0.33333333 0.66666667]
# [ 0.25 0.33333333 0.41666667]
# [ 0.28571429 0.33333333 0.38095238]]
Now your rows will sum to 1.
Solution 3
I think this should work,
a = numpy.arange(0,27.,3).reshape(3,3)
a /= a.sum(axis=1)[:,numpy.newaxis]
Solution 4
In case you are trying to normalize each row such that its magnitude is one (i.e. a row's unit length is one or the sum of the square of each element in a row is one):
import numpy as np
a = np.arange(0,27,3).reshape(3,3)
result = a / np.linalg.norm(a, axis=-1)[:, np.newaxis]
# array([[ 0. , 0.4472136 , 0.89442719],
# [ 0.42426407, 0.56568542, 0.70710678],
# [ 0.49153915, 0.57346234, 0.65538554]])
Verifying:
np.sum( result**2, axis=-1 )
# array([ 1., 1., 1.])
Solution 5
I think you can normalize the row elements sum to 1 by this:
new_matrix = a / a.sum(axis=1, keepdims=1).
And the column normalization can be done with new_matrix = a / a.sum(axis=0, keepdims=1). Hope this can hep.
Related videos on Youtube
Aufwind
Updated on October 14, 2021Comments
-
Aufwind over 2 years
Given a 3 times 3 numpy array
a = numpy.arange(0,27,3).reshape(3,3) # array([[ 0, 3, 6], # [ 9, 12, 15], # [18, 21, 24]])To normalize the rows of the 2-dimensional array I thought of
row_sums = a.sum(axis=1) # array([ 9, 36, 63]) new_matrix = numpy.zeros((3,3)) for i, (row, row_sum) in enumerate(zip(a, row_sums)): new_matrix[i,:] = row / row_sumThere must be a better way, isn't there?
Perhaps to clearify: By normalizing I mean, the sum of the entrys per row must be one. But I think that will be clear to most people.
-
coldfix almost 9 yearsCareful, "normalize" usually means the square sum of components is one. Your definition will hardly be clear to most people;)
-
 Bálint Sass over 3 years@coldfix speaks about
Bálint Sass over 3 years@coldfix speaks aboutL2norm and considers it as most common (which may be true) while Aufwind usesL1norm which is also a norm indeed.
-
-
 wim over 12 yearsgood. note the change of dtype to arange, by appending decimal point to 27.
wim over 12 yearsgood. note the change of dtype to arange, by appending decimal point to 27. -
Ztyx almost 10 yearsAxis doesn't seem to be a parameter to np.linalg.norm (anymore?).
-
dpb over 9 yearsnotably this corresponds to the l2 norm (where as rows summing to 1 corresponds to the l1 norm)
-
ali_m about 9 yearsThis can be simplified even further using
a.sum(axis=1, keepdims=True)to keep the singleton column dimension, which you can then broadcast along without having to usenp.newaxis. -
asdf about 9 yearswhat if any of the row_sums is zero?
-
ali_m about 9 years@asdf ...well in that case normalizing by the row sum doesn't really make much sense!
-
coldfix almost 9 yearsThis is the correct answer for the question as stated above - but if a normalization in the usual sense is desired, use
np.linalg.norminstead ofa.sum! -
Paul almost 9 yearsis this preferred to
row_sums.reshape(3,1)? -
nos almost 8 yearsIt's not as robust since the row sum may be 0.
-
XY.W over 7 yearsIf a vector is normalized, it should have a unit norm, using a / row_sums[:, numpy.newaxis] really doesn't guarantee a unit norm.
-
Bi Rico over 7 years@XY.W There are many definitions of "unit norm", take a look at the ord argument to numpy's norm function. Ord 1 norms are often useful and the OP asked specifically about normalizing with respect to this norm, but you can of course replace the denominator with the most appropriate norm for your application.
-
 Mona Jalal over 6 yearsIs this the same as MinMaxNorm or what is the name of this normalization?
Mona Jalal over 6 yearsIs this the same as MinMaxNorm or what is the name of this normalization? -
JEM_Mosig over 4 yearsThis also has the advantage that it works on sparse arrays that would not fit into memory as dense arrays.
-
Johannes Ackermann about 3 yearsThis is equivalent to
new_matrix = a / row_sums[:, None], asNonecan be used as a shorthand fornp.newaxis. -
qwr about 2 yearsthis answer is incomplete without how you computed
row_sums -
qwr about 2 yearsThis computes the norm and does not normalize the matrix
-
qwr about 2 yearsis this using python's
map? won't builtin numpy functions be much faster? -
qwr about 2 yearstoo inefficient. you turned a simple sum over all elements into a big (sparse) matrix multiplication
-
 Maciek about 2 yearsIt is in the original question:
Maciek about 2 yearsIt is in the original question:row_sums = a.sum(axis=1)