pandas dataframe columns scaling with sklearn
Solution 1
I am not sure if previous versions of pandas prevented this but now the following snippet works perfectly for me and produces exactly what you want without having to use apply
>>> import pandas as pd
>>> from sklearn.preprocessing import MinMaxScaler
>>> scaler = MinMaxScaler()
>>> dfTest = pd.DataFrame({'A':[14.00,90.20,90.95,96.27,91.21],
'B':[103.02,107.26,110.35,114.23,114.68],
'C':['big','small','big','small','small']})
>>> dfTest[['A', 'B']] = scaler.fit_transform(dfTest[['A', 'B']])
>>> dfTest
A B C
0 0.000000 0.000000 big
1 0.926219 0.363636 small
2 0.935335 0.628645 big
3 1.000000 0.961407 small
4 0.938495 1.000000 small
Solution 2
Like this?
dfTest = pd.DataFrame({
'A':[14.00,90.20,90.95,96.27,91.21],
'B':[103.02,107.26,110.35,114.23,114.68],
'C':['big','small','big','small','small']
})
dfTest[['A','B']] = dfTest[['A','B']].apply(
lambda x: MinMaxScaler().fit_transform(x))
dfTest
A B C
0 0.000000 0.000000 big
1 0.926219 0.363636 small
2 0.935335 0.628645 big
3 1.000000 0.961407 small
4 0.938495 1.000000 small
Solution 3
df = pd.DataFrame(scale.fit_transform(df.values), columns=df.columns, index=df.index)
This should work without depreciation warnings.
Solution 4
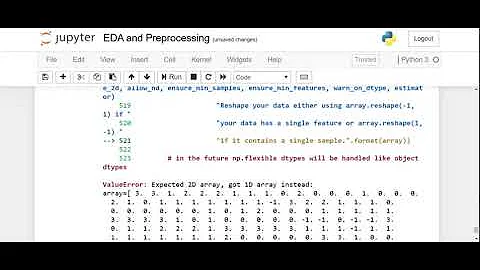
As it is being mentioned in pir's comment - the .apply(lambda el: scale.fit_transform(el)) method will produce the following warning:
DeprecationWarning: Passing 1d arrays as data is deprecated in 0.17 and will raise ValueError in 0.19. Reshape your data either using X.reshape(-1, 1) if your data has a single feature or X.reshape(1, -1) if it contains a single sample.
Converting your columns to numpy arrays should do the job (I prefer StandardScaler):
from sklearn.preprocessing import StandardScaler
scale = StandardScaler()
dfTest[['A','B','C']] = scale.fit_transform(dfTest[['A','B','C']].as_matrix())
-- Edit Nov 2018 (Tested for pandas 0.23.4)--
As Rob Murray mentions in the comments, in the current (v0.23.4) version of pandas .as_matrix() returns FutureWarning. Therefore, it should be replaced by .values:
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit_transform(dfTest[['A','B']].values)
-- Edit May 2019 (Tested for pandas 0.24.2)--
As joelostblom mentions in the comments, "Since 0.24.0, it is recommended to use .to_numpy() instead of .values."
Updated example:
import pandas as pd
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
dfTest = pd.DataFrame({
'A':[14.00,90.20,90.95,96.27,91.21],
'B':[103.02,107.26,110.35,114.23,114.68],
'C':['big','small','big','small','small']
})
dfTest[['A', 'B']] = scaler.fit_transform(dfTest[['A','B']].to_numpy())
dfTest
A B C
0 -1.995290 -1.571117 big
1 0.436356 -0.603995 small
2 0.460289 0.100818 big
3 0.630058 0.985826 small
4 0.468586 1.088469 small
Solution 5
You can do it using pandas only:
In [235]:
dfTest = pd.DataFrame({'A':[14.00,90.20,90.95,96.27,91.21],'B':[103.02,107.26,110.35,114.23,114.68], 'C':['big','small','big','small','small']})
df = dfTest[['A', 'B']]
df_norm = (df - df.min()) / (df.max() - df.min())
print df_norm
print pd.concat((df_norm, dfTest.C),1)
A B
0 0.000000 0.000000
1 0.926219 0.363636
2 0.935335 0.628645
3 1.000000 0.961407
4 0.938495 1.000000
A B C
0 0.000000 0.000000 big
1 0.926219 0.363636 small
2 0.935335 0.628645 big
3 1.000000 0.961407 small
4 0.938495 1.000000 small
Related videos on Youtube
flyingmeatball
Updated on March 03, 2022Comments
-
 flyingmeatball about 2 years
flyingmeatball about 2 yearsI have a pandas dataframe with mixed type columns, and I'd like to apply sklearn's min_max_scaler to some of the columns. Ideally, I'd like to do these transformations in place, but haven't figured out a way to do that yet. I've written the following code that works:
import pandas as pd import numpy as np from sklearn import preprocessing scaler = preprocessing.MinMaxScaler() dfTest = pd.DataFrame({'A':[14.00,90.20,90.95,96.27,91.21],'B':[103.02,107.26,110.35,114.23,114.68], 'C':['big','small','big','small','small']}) min_max_scaler = preprocessing.MinMaxScaler() def scaleColumns(df, cols_to_scale): for col in cols_to_scale: df[col] = pd.DataFrame(min_max_scaler.fit_transform(pd.DataFrame(dfTest[col])),columns=[col]) return df dfTest A B C 0 14.00 103.02 big 1 90.20 107.26 small 2 90.95 110.35 big 3 96.27 114.23 small 4 91.21 114.68 small scaled_df = scaleColumns(dfTest,['A','B']) scaled_df A B C 0 0.000000 0.000000 big 1 0.926219 0.363636 small 2 0.935335 0.628645 big 3 1.000000 0.961407 small 4 0.938495 1.000000 smallI'm curious if this is the preferred/most efficient way to do this transformation. Is there a way I could use df.apply that would be better?
I'm also surprised I can't get the following code to work:
bad_output = min_max_scaler.fit_transform(dfTest['A'])If I pass an entire dataframe to the scaler it works:
dfTest2 = dfTest.drop('C', axis = 1) good_output = min_max_scaler.fit_transform(dfTest2) good_outputI'm confused why passing a series to the scaler fails. In my full working code above I had hoped to just pass a series to the scaler then set the dataframe column = to the scaled series.
-
EdChum almost 10 yearsDoes it work if you do this
bad_output = min_max_scaler.fit_transform(dfTest['A'].values)? accessing thevaluesattribute returns a numpy array, for some reason sometimes the scikit learn api will correctly call the right method that makes pandas returns a numpy array and sometimes it doesn't. -
Fred Foo almost 10 yearsPandas' dataframes are quite complicated objects with conventions that do not match scikit-learn's conventions. If you convert everything to NumPy arrays, scikit-learn gets a lot easier to work with.
-
 flyingmeatball almost 10 years@edChum -
flyingmeatball almost 10 years@edChum -bad_output = in_max_scaler.fit_transform(dfTest['A'].values)did not work either. @larsmans - yeah I had thought about going down this route, it just seems like a hassle. I don't know if it is a bug or not that Pandas can pass a full dataframe to a sklearn function, but not a series. My understanding of a dataframe was that it is a dict of series. Reading in the "Python for Data Analysis" book, it states that pandas is built on top of numpy to make it easy to use in NumPy-centric applicatations.
-
-
 flyingmeatball almost 10 yearsI know that I can do it just in pandas, but I may want to eventually apply a different sklearn method that isn't as easy to write myself. I'm more interested in figuring out why applying to a series doesn't work as I expected than I am in coming up with a strictly simpler solution. My next step will be to run a RandomForestRegressor, and I want to make sure I understand how Pandas and sklearn work together.
flyingmeatball almost 10 yearsI know that I can do it just in pandas, but I may want to eventually apply a different sklearn method that isn't as easy to write myself. I'm more interested in figuring out why applying to a series doesn't work as I expected than I am in coming up with a strictly simpler solution. My next step will be to run a RandomForestRegressor, and I want to make sure I understand how Pandas and sklearn work together. -
pir about 8 yearsI get a bunch of DeprecationWarnings when I run this script. How should it be updated?
-
 AJP over 7 yearsSee @LetsPlayYahtzee's answer below
AJP over 7 yearsSee @LetsPlayYahtzee's answer below -
 citynorman over 6 yearsNeat! A more generalized version
citynorman over 6 yearsNeat! A more generalized versiondf[df.columns] = scaler.fit_transform(df[df.columns]) -
Rajesh Mappu over 6 yearsI know this is a delayed comment from original date, but why is there two square brackets in dfTest[['A', 'B']]? I can see it doesn't work with single bracket, but couldn't understand the reason.
-
 ken about 6 years@RajeshThevar The outer brackets are pandas' typical selector brackets, telling pandas to select a column from the dataframe. The inner brackets indicate a list. You're passing a list to the pandas selector. If you just use single brackets - with one column name followed by another, separated by a comma - pandas interprets this as if you're trying to select a column from a dataframe with multi-level columns (a MultiIndex) and will throw a keyerror.
ken about 6 years@RajeshThevar The outer brackets are pandas' typical selector brackets, telling pandas to select a column from the dataframe. The inner brackets indicate a list. You're passing a list to the pandas selector. If you just use single brackets - with one column name followed by another, separated by a comma - pandas interprets this as if you're trying to select a column from a dataframe with multi-level columns (a MultiIndex) and will throw a keyerror. -
 LetsPlayYahtzee about 6 yearsto add to @ken's answer if you want to see exactly how pandas implements this indexing logic and why a tuple of values would be interpreted differently than a list you can look at how DataFrames implement the
LetsPlayYahtzee about 6 yearsto add to @ken's answer if you want to see exactly how pandas implements this indexing logic and why a tuple of values would be interpreted differently than a list you can look at how DataFrames implement the__getitem__method. Specifically you can open you ipython and dopd.DataFrame.__getitem__??; after you import pandas as pd of course ;) -
Adam Stewart almost 6 yearsThis works great, however when I try to do
scaler.fit()andscaler.transform()on two separate lines I'm getting the dreadedSettingWithCopyWarning. Anyone have any idea why? -
David J. over 5 yearsA practical note: for those using train/test data splits, you'll want to only fit on your training data, not your testing data.
-
 satsumas over 5 years
satsumas over 5 yearsdf[df.columns] = scaler.fit_transform(df[df.columns])-- perfect @citynorman! -
 Asclepius over 5 yearsThis answer is dangerous because
Asclepius over 5 yearsThis answer is dangerous becausedf.max() - df.min()can be 0, leading to an exception. Moreover,df.min()is computed twice which is inefficient. Note thatdf.ptp()is equivalent todf.max() - df.min(). -
Alexandre V. over 5 yearsA simpler version: dfTest[['A','B']] = dfTest[['A','B']].apply(MinMaxScaler().fit_transform)
-
 Rob Murray over 5 yearsuse
Rob Murray over 5 yearsuse.valuesin place of.as_matrix()asas_matrix()now gives aFutureWarning. -
 intotecho over 5 yearsTo scale all but the timestamps column, combine with
intotecho over 5 yearsTo scale all but the timestamps column, combine withcolumns =df.columns.drop('timestamps') df[df.columns] = scaler.fit_transform(df[df.columns] -
 compguy24 about 5 years@DavidJ. can you provide a reference for why?
compguy24 about 5 years@DavidJ. can you provide a reference for why? -
 joelostblom almost 5 yearsSince
joelostblom almost 5 yearsSince0.24.0, it is recommended to use.to_numpy()instead of.values. -
 JolonB almost 4 yearsCorrection of @intotecho's comment. You should do
JolonB almost 4 yearsCorrection of @intotecho's comment. You should docolumns = df.columns.drop('timestamps')anddf[columns] = scaler.fit_transform(df[columns]). It should becolumnsin the square brackets, notdf.columns -
HFX over 3 yearsHow to inverse a value in this way?
-
 shcrela about 3 yearsor
shcrela about 3 yearsordf[df.columns] = scale.fit_transform(df) -
Ammanuel over 2 yearsWorks perfectly! I was trying to figure out how to retain the column names, this helped.
-
 ssp over 2 yearsCan use
ssp over 2 yearsCan usedf[:] = scaler.fit_transform(df)if scaling the entire dataset. -
ICW almost 2 yearsthis will instantiate a new MinMaxScaler per row not sure if it matters though