Python chi square goodness of fit test to get the best distribution
Solution 1
Python chi square goodness of fit test (https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.chisquare.html) mentions that "“Delta degrees of freedom”: adjustment to the degrees of freedom for the p-value. The p-value is computed using a chi-squared distribution with k - 1 - ddof degrees of freedom, where k is the number of observed frequencies. The default value of ddof is 0."
Hence your code should be corrected as follows.
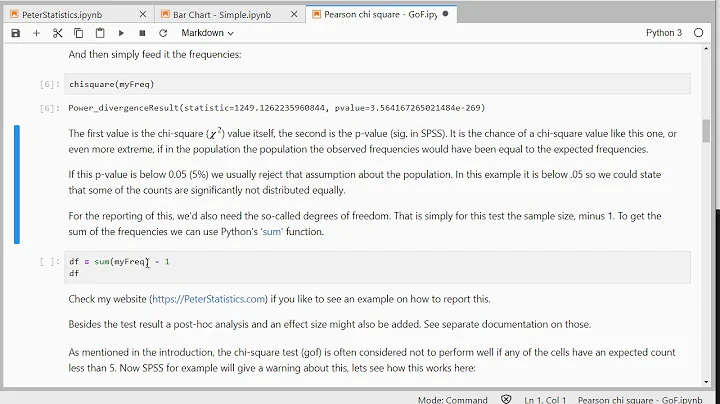
c , p = st.chisquare(observed_values, expected_values, ddof=len(param))
Solution 2
The Pareto function you are using to draw the random number is not the same as the one you are using to fit the data.
The first one is from numpy and they state
Draw samples from a Pareto II or Lomax distribution with specified shape.
The Lomax or Pareto II distribution is a shifted Pareto distribution. The classical Pareto distribution can be obtained from the Lomax distribution by adding 1 and multiplying by the scale parameter m.
The pareto function you use to fit is the one from Scipy and I guess they use a different definition:
The probability density above is defined in the “standardized” form. To shift and/or scale the distribution use the loc and scale parameters.
Related videos on Youtube
Pasindu Tennage
Updated on September 16, 2022Comments
-
 Pasindu Tennage over 1 year
Pasindu Tennage over 1 yearGiven a set of data values, I'm trying to get the best theoretical distribution that describes the data well. I came up with the following python code after days of research.
import numpy as np import csv import pandas as pd import scipy.stats as st import math import sys import matplotlib matplotlib.use('Agg') import matplotlib.pyplot as plt def fit_to_all_distributions(data): dist_names = ['fatiguelife', 'invgauss', 'johnsonsu', 'johnsonsb', 'lognorm', 'norminvgauss', 'powerlognorm', 'exponweib','genextreme', 'pareto'] params = {} for dist_name in dist_names: try: dist = getattr(st, dist_name) param = dist.fit(data) params[dist_name] = param except Exception: print("Error occurred in fitting") params[dist_name] = "Error" return params def get_best_distribution_using_chisquared_test(data, params): histo, bin_edges = np.histogram(data, bins='auto', normed=False) number_of_bins = len(bin_edges) - 1 observed_values = histo dist_names = ['fatiguelife', 'invgauss', 'johnsonsu', 'johnsonsb', 'lognorm', 'norminvgauss', 'powerlognorm', 'exponweib','genextreme', 'pareto'] dist_results = [] for dist_name in dist_names: param = params[dist_name] if (param != "Error"): # Applying the SSE test arg = param[:-2] loc = param[-2] scale = param[-1] cdf = getattr(st, dist_name).cdf(bin_edges, loc=loc, scale=scale, *arg) expected_values = len(data) * np.diff(cdf) c , p = st.chisquare(observed_values, expected_values, ddof=number_of_bins-len(param)) dist_results.append([dist_name, c, p]) # select the best fitted distribution best_dist, best_c, best_p = None, sys.maxsize, 0 for item in dist_results: name = item[0] c = item[1] p = item[2] if (not math.isnan(c)): if (c < best_c): best_c = c best_dist = name best_p = p # print the name of the best fit and its p value print("Best fitting distribution: " + str(best_dist)) print("Best c value: " + str(best_c)) print("Best p value: " + str(best_p)) print("Parameters for the best fit: " + str(params[best_dist])) return best_dist, best_c, params[best_dist], dist_resultsThen I test this code by,
a, m = 3., 2. values = (np.random.pareto(a, 1000) + 1) * m data = pd.Series(values) params = fit_to_all_distributions(data) best_dist_chi, best_chi, params_chi, dist_results_chi = get_best_distribution_using_chisquared_test(values, params)Since the data points are generated using Pareto distribution, it should return pareto as the best fitting distribution with a sufficiently large p value (p>0.05).
But this is what I get as output.
Best fitting distribution: genextreme Best c value: 106.46087793622216 Best p value: 7.626303538461713e-24 Parameters for the best fit: (-0.7664124294696955, 2.3217378846757164, 0.3711562696710188)Is there anything wrong with my implementation of Chi Squared goodness of fit test?