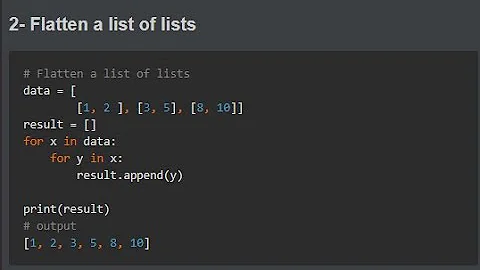
How to flatten a list of lists?
Solution 1
I expect that unlist(foolist) will help you. It has an option recursive which is TRUE by default.
So unlist(foolist, recursive = FALSE) will return the list of the documents, and then you can combine them by:
do.call(c, unlist(foolist, recursive=FALSE))
do.call just applies the function c to the elements of the obtained list
Solution 2
Here's a more general solution for when lists are nested multiple times and the amount of nesting differs between elements of the lists:
flattenlist <- function(x){
morelists <- sapply(x, function(xprime) class(xprime)[1]=="list")
out <- c(x[!morelists], unlist(x[morelists], recursive=FALSE))
if(sum(morelists)){
Recall(out)
}else{
return(out)
}
}
Solution 3
Here's another method that worked for my list of lists.
df <- as.data.frame(do.call(rbind, lapply(foolist, as.data.frame)))
Or take a look at new functions in tidyr which work well.
rectangle a nested list into a tidy tibble
lst <- list(
list(
age = 23,
gender = "Male",
city = "Sydney"
),
list(
age = 21,
gender = "Female",
city = "Cairns"
)
)
tib <- tibble(lst) %>%
unnest_wider(lst)
df <- as.data.frame(tib)
Related videos on Youtube
dnagirl
Updated on May 30, 2021Comments
-
dnagirl almost 3 years
The
tmpackage extendscso that, if given a set ofPlainTextDocuments it automatically creates aCorpus. Unfortunately, it appears that eachPlainTextDocumentmust be specified separately.e.g. if I had:
foolist <- list(a, b, c); # where a,b,c are PlainTextDocument objectsI'd do this to get a
Corpus:foocorpus <- c(foolist[[1]], foolist[[2]], foolist[[3]]);I have a list of lists of
'PlainTextDocuments that looks like this:> str(sectioned) List of 154 $ :List of 6 ..$ :Classes 'PlainTextDocument', 'TextDocument', 'character' atomic [1:1] Developing assessment models Developing models .. .. ..- attr(*, "Author")= chr "John Smith" .. .. ..- attr(*, "DateTimeStamp")= POSIXlt[1:1], format: "2013-04-30 12:03:49" .. .. ..- attr(*, "Description")= chr(0) .. .. ..- attr(*, "Heading")= chr "Research Focus" .. .. ..- attr(*, "ID")= chr(0) .. .. ..- attr(*, "Language")= chr(0) .. .. ..- attr(*, "LocalMetaData")=List of 4 .. .. .. ..$ foo : chr "bar" .. .. .. ..$ classification: chr "Technician" .. .. .. ..$ team : chr "" .. .. .. ..$ supervisor : chr "Bill Jones" .. .. ..- attr(*, "Origin")= chr "Smith-John_e.txt" #etc., all sublists have 6 elementsSo, to get all my
PlainTextDocuments into aCorpus, this would work:sectioned.Corpus <- c(sectioned[[1]][[1]], sectioned[[1]][[2]], ..., sectioned[[154]][[6]])Can anyone suggest an easier way, please?
ETA:
foo<-unlist(foolist, recursive=FALSE)produces a flat list of PlainTextDocuments, which still leaves me with the problem of feeding a list element by element toc -
Michael about 7 yearsjust to make this a bit more comprehensible I'll just point out that identifying lists using class(xprime)[1]=="list") is necessary (rather than using is.list) when your nested objects are of classes that inherit from lists (i.e. note that is.list(data.frame(3)) evaluates to TRUE)
-
Michael about 7 yearsalso note that this doesn't retain the order of the original structure
-
 Megatron over 5 yearsAlso consider using
Megatron over 5 yearsAlso consider usingNCmisc::Unlist()for unlisting beyond the first level. -
 Mihai over 4 yearsMichael, how would you go about retaining the order of the original structure?
Mihai over 4 yearsMichael, how would you go about retaining the order of the original structure? -
Michael about 3 yearsprobably just replace
out <- c(with anmapplystatement that takesxandmorelistsas arguments then unlists only for elements wheremorelistsis TRUE -
Michael about 3 yearsor replace the first two lines with a single
lapplythat combines testing and unlisting -
Simone over 2 yearsThanks for sharing.
unnest_widerfrom the tidyr package worked perfectly. The other code you posted gave me an error message "Error in match.names(clabs, names(xi)) : names do not match previous names" --> The data I am working with is downloaded as a JSON from Facebook. It is highly nested. Not all lists in the list are equally long. The data structure also varies between downloaded files. -
 Zeus over 2 years@Simone, I think you should post a new question with a simple replication of your data, then some can try to help you. SO doesn't like questions within questions
Zeus over 2 years@Simone, I think you should post a new question with a simple replication of your data, then some can try to help you. SO doesn't like questions within questions -
Simone over 2 yearsI did. Wanted to point out that there are "normally" nested lists and highly nested lists. For the latter the tidyr package is useful. --> hence the comment and not a new question.
-
 shosaco over 2 yearsVery useful: My usecase is
shosaco over 2 yearsVery useful: My usecase ismap(some_nested_list, flattenlist) %>% bind_rows()to produce a tibble. -
choabf about 2 years@zx8754 for the very similar large list, after importing a json file, I'm applying the same code, but I get the error "unlist arguments imply differing number of rows: 1, 0", which I understand, but i thought the code deals with lists of different levels. Any ideas? Beginner R user here... Thanks in advance for any help!
-
DrDom about 2 years@choabf, I advise you to create a new question, where put an example of your data and code you used.