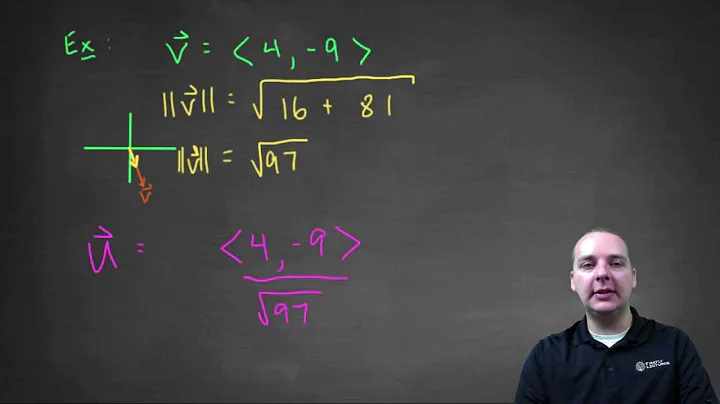How to normalize a NumPy array to a unit vector?
Solution 1
If you're using scikit-learn you can use sklearn.preprocessing.normalize:
import numpy as np
from sklearn.preprocessing import normalize
x = np.random.rand(1000)*10
norm1 = x / np.linalg.norm(x)
norm2 = normalize(x[:,np.newaxis], axis=0).ravel()
print np.all(norm1 == norm2)
# True
Solution 2
I agree that it would be nice if such a function were part of the included libraries. But it isn't, as far as I know. So here is a version for arbitrary axes that gives optimal performance.
import numpy as np
def normalized(a, axis=-1, order=2):
l2 = np.atleast_1d(np.linalg.norm(a, order, axis))
l2[l2==0] = 1
return a / np.expand_dims(l2, axis)
A = np.random.randn(3,3,3)
print(normalized(A,0))
print(normalized(A,1))
print(normalized(A,2))
print(normalized(np.arange(3)[:,None]))
print(normalized(np.arange(3)))
Solution 3
This might also work for you
import numpy as np
normalized_v = v / np.sqrt(np.sum(v**2))
but fails when v has length 0.
In that case, introducing a small constant to prevent the zero division solves this.
Solution 4
To avoid zero division I use eps, but that's maybe not great.
def normalize(v):
norm=np.linalg.norm(v)
if norm==0:
norm=np.finfo(v.dtype).eps
return v/norm
Solution 5
If you have multidimensional data and want each axis normalized to its max or its sum:
def normalize(_d, to_sum=True, copy=True):
# d is a (n x dimension) np array
d = _d if not copy else np.copy(_d)
d -= np.min(d, axis=0)
d /= (np.sum(d, axis=0) if to_sum else np.ptp(d, axis=0))
return d
Uses numpys peak to peak function.
a = np.random.random((5, 3))
b = normalize(a, copy=False)
b.sum(axis=0) # array([1., 1., 1.]), the rows sum to 1
c = normalize(a, to_sum=False, copy=False)
c.max(axis=0) # array([1., 1., 1.]), the max of each row is 1
Related videos on Youtube
Donbeo
Updated on May 03, 2022Comments
-
Donbeo about 2 years
I would like to convert a NumPy array to a unit vector. More specifically, I am looking for an equivalent version of this normalisation function:
def normalize(v): norm = np.linalg.norm(v) if norm == 0: return v return v / normThis function handles the situation where vector
vhas the norm value of 0.Is there any similar functions provided in
sklearnornumpy?-
ali_m over 10 yearsWhat's wrong with what you've written?
-
 Hooked over 10 yearsIf this is really a concern, you should check for norm < epsilon, where epsilon is a small tolerance. In addition, I wouldn't silently pass back a norm zero vector, I would
Hooked over 10 yearsIf this is really a concern, you should check for norm < epsilon, where epsilon is a small tolerance. In addition, I wouldn't silently pass back a norm zero vector, I wouldraisean exception! -
Donbeo over 10 yearsmy function works but I would like to know if there is something inside the python's more common library. I am writing different machine learning functions and I would like to avoid to define too much new functions to make the code more clear and readable
-
Bill over 5 yearsI did a few quick tests and I found that
x/np.linalg.norm(x)was not much slower (about 15-20%) thanx/np.sqrt((x**2).sum())in numpy 1.15.1 on a CPU.
-
-
Donbeo over 10 yearsThanks for the answer but are you sure that sklearn.preprocessing.normalize works also with vector of shape=(n,) or (n,1) ? I am having some problems with this library
-
ali_m over 10 years
normalizerequires a 2D input. You can pass theaxis=argument to specify whether you want to apply the normalization across the rows or columns of your input array. -
Donbeo over 10 yearsI did not deeply test the ali_m solution but in some simple case it seems to be working. Are there situtions where your function does better?
-
Eelco Hoogendoorn over 10 yearsI don't know; but it works over arbitrary axes, and we have explicit control over what happens for length 0 vectors.
-
 Neil G over 9 yearsVery nice! This should be in numpy — although order should probably come before axis in my opinion.
Neil G over 9 yearsVery nice! This should be in numpy — although order should probably come before axis in my opinion. -
Henry Thornton almost 9 years@EelcoHoogendoorn Curious to understand why order=2 chosen over others?
-
Eelco Hoogendoorn almost 9 yearsBecause the Euclidian/pythagoran norm happens to be the most frequently used one; wouldn't you agree?
-
Ash over 8 yearsNote that the 'norm' argument of the normalize function can be either 'l1' or 'l2' and the default is 'l2'. If you want your vector's sum to be 1 (e.g. a probability distribution) you should use norm='l1' in the normalize function.
-
bendl almost 7 yearsPretty late, but I think it's worth mentioning that this is exactly why it is discouraged to use lowercase 'L' as a variable name... in my typeface 'l2' is indistinguishable from '12'
-
pasbi about 6 yearsnormalizing
[inf, 1, 2]yields[nan, 0, 0], but shouldn't it be[1, 0, 0]? -
pasbi about 6 yearsnormalizing
[inf, 1, 2]yields[nan, 0, 0], but shouldn't it be[1, 0, 0]? -
Eelco Hoogendoorn about 6 yearsIf you'd like to endow the fp-inf symbol with such semantics, sure, but thatd be kinda nonstandard. The fp standard is full of quirks anyway but I think having such a function do anything but standard fp logic by default would just be confusing.
-
 Spenhouet almost 6 yearsShouldn't the normalized array sum up to 1 (at least I would expect it to do)? Just tested this implementation with
Spenhouet almost 6 yearsShouldn't the normalized array sum up to 1 (at least I would expect it to do)? Just tested this implementation with[5,5]what yields[0.70710678, 0.70710678]what in sum is about1.41. Doesn't sound right to me. -
Eelco Hoogendoorn almost 6 yearsLook up the concept of the order of a norm. What you want is the 1-norm, which you can get by setting the order kwarg to 1.
-
Omid almost 6 yearsAlso note that
np.linalg.norm(x)calculates 'l2' norm by default. If you want your vector's sum to be 1 you should usenp.linalg.norm(x, ord=1) -
Milso about 4 yearsWatch out if all values are the same in the original matrix, then ptp would be 0. Division by 0 will return nan.
-
 Ramin Melikov about 4 yearsNote: x must be
Ramin Melikov about 4 yearsNote: x must bendarrayfor it to work with thenormalize()function. Otherwise it can be alist. -
 crypdick over 3 yearsThis does a different type of transform. The OP wanted to scale the magnitude of the vector so that each vector has a length of 1; MinMaxScaler individually scales each column independently to be within a certain range.
crypdick over 3 yearsThis does a different type of transform. The OP wanted to scale the magnitude of the vector so that each vector has a length of 1; MinMaxScaler individually scales each column independently to be within a certain range. -
 crypdick over 3 yearsThese output arrays do not have unit norm. Subtracting the mean and giving the samples unit variance does not produce unit vectors.
crypdick over 3 yearsThese output arrays do not have unit norm. Subtracting the mean and giving the samples unit variance does not produce unit vectors. -
 anon01 about 3 years@bendl I think that's exactly why it's encouraged to use a better typeface
anon01 about 3 years@bendl I think that's exactly why it's encouraged to use a better typeface -
 Alessandro Muzzi over 2 yearsSome time has passed but the answer is no,
Alessandro Muzzi over 2 yearsSome time has passed but the answer is no,[nan, 0, 0]is correct since the norm isinfandinf/infis an indeterminate form because<everything>/infis0but is also true thatinf/<everything>isinf, soinf/infcannot be determined. -
John Henckel over 2 yearsRegarding choice of variable name
l2, most python users are not software engineers. They are mathematicians and scientists first, and their code tends to reflect that culture, where using single letters for variables is customary. I agree though. Using lowercaselor uppercaseOas a variable names should definitely be avoided. Trust me, I fix software bugs for a living. -
NerdOnTour over 2 yearsIs there a reason for you to use the L1-norm? The OP seems to ask for L2-normalization.
-
user111950 about 2 yearsI often use this trick: x_normalised = x / (norm+(norm==0)) so in all cases where the norm is zero, you just divide by one.
-
 Eduard Feicho about 2 yearshm yeah should have been l2 norm
Eduard Feicho about 2 yearshm yeah should have been l2 norm -
scign almost 2 years@pasbi then what should
[inf, 3, inf]yield?[1, 0, 1]or[0.5, 0, 0.5]or something else? I'd say[nan, 0, nan]would be the right output so the user can then fill thenanvalues with your chosen filler to force the expectation. -
pasbi almost 2 years@scign I guess there's no right or wrong here, just more or less useful. I agree that your proposal is probably more useful in general. It's also more IEEE 754-conform (as
∞/∞=nan). I don't remember my use case from four years ago, I presume it was something very special.