Date axis in heatmap seaborn
Solution 1
You have to use strftime function for your date series of dataframe to plot xtick labels correctly:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from datetime import datetime, timedelta
import random
dates = [datetime.today() - timedelta(days=x * random.getrandbits(1)) for x in xrange(25)]
df = pd.DataFrame({'depth': [0.1,0.05, 0.01, 0.005, 0.001, 0.1, 0.05, 0.01, 0.005, 0.001, 0.1, 0.05, 0.01, 0.005, 0.001, 0.1, 0.05, 0.01, 0.005, 0.001, 0.1, 0.05, 0.01, 0.005, 0.001],\
'date': dates,\
'value': [-4.1808639999999997, -9.1753490000000006, -11.408113999999999, -10.50245, -8.0274750000000008, -0.72260200000000008, -6.9963940000000004, -10.536339999999999, -9.5440649999999998, -7.1964070000000007, -0.39225599999999999, -6.6216390000000001, -9.5518009999999993, -9.2924690000000005, -6.7605589999999998, -0.65214700000000003, -6.8852289999999989, -9.4557760000000002, -8.9364629999999998, -6.4736289999999999, -0.96481800000000006, -6.051482, -9.7846860000000007, -8.5710630000000005, -6.1461209999999999]})
pivot = df.pivot(index='depth', columns='date', values='value')
sns.set()
ax = sns.heatmap(pivot)
ax.set_xticklabels(df['date'].dt.strftime('%d-%m-%Y'))
plt.xticks(rotation=-90)
plt.show()
Solution 2
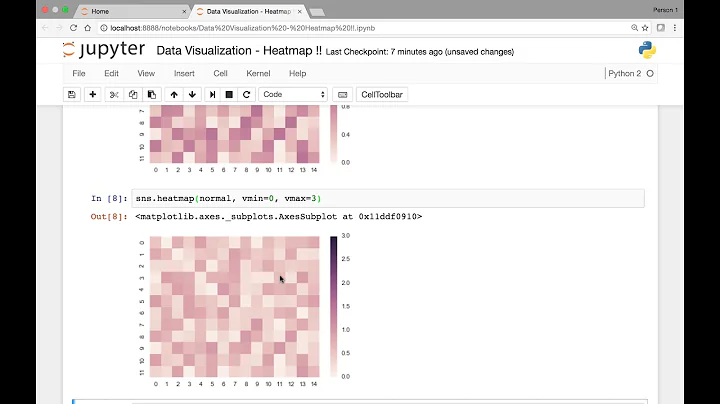
Example with standard heatmap datetime labels
import pandas as pd
import seaborn as sns
dates = pd.date_range('2019-01-01', '2020-12-01')
df = pd.DataFrame(np.random.randint(0, 100, size=(len(dates), 4)), index=dates)
sns.heatmap(df)
We can create some helper classes/functions to get to some better looking labels and placement. AxTransformer enables conversion from data coordinates to tick locations, set_date_ticks allows custom date ranges to be applied to plots.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from collections.abc import Iterable
from sklearn import linear_model
class AxTransformer:
def __init__(self, datetime_vals=False):
self.datetime_vals = datetime_vals
self.lr = linear_model.LinearRegression()
return
def process_tick_vals(self, tick_vals):
if not isinstance(tick_vals, Iterable) or isinstance(tick_vals, str):
tick_vals = [tick_vals]
if self.datetime_vals == True:
tick_vals = pd.to_datetime(tick_vals).astype(int).values
tick_vals = np.array(tick_vals)
return tick_vals
def fit(self, ax, axis='x'):
axis = getattr(ax, f'get_{axis}axis')()
tick_locs = axis.get_ticklocs()
tick_vals = self.process_tick_vals([label._text for label in axis.get_ticklabels()])
self.lr.fit(tick_vals.reshape(-1, 1), tick_locs)
return
def transform(self, tick_vals):
tick_vals = self.process_tick_vals(tick_vals)
tick_locs = self.lr.predict(np.array(tick_vals).reshape(-1, 1))
return tick_locs
def set_date_ticks(ax, start_date, end_date, axis='y', date_format='%Y-%m-%d', **date_range_kwargs):
dt_rng = pd.date_range(start_date, end_date, **date_range_kwargs)
ax_transformer = AxTransformer(datetime_vals=True)
ax_transformer.fit(ax, axis=axis)
getattr(ax, f'set_{axis}ticks')(ax_transformer.transform(dt_rng))
getattr(ax, f'set_{axis}ticklabels')(dt_rng.strftime(date_format))
ax.tick_params(axis=axis, which='both', bottom=True, top=False, labelbottom=True)
return ax
These provide us a lot of flexibility, e.g.
fig, ax = plt.subplots(dpi=150)
sns.heatmap(df, ax=ax)
set_date_ticks(ax, '2019-01-01', '2020-12-01', freq='3MS')
or if you really want to get weird you can do stuff like
fig, ax = plt.subplots(dpi=150)
sns.heatmap(df, ax=ax)
set_date_ticks(ax, '2019-06-01', '2020-06-01', freq='2MS', date_format='%b `%y')
For your specific example you'll have to pass axis='x' to set_date_ticks
Solution 3
- First, the
'date'column must be converted to adatetime dtypewithpandas.to_datetime - If the desired result is to only have the dates (without time), then the easiest solution is to use the
.dtaccessor to extract the.datecomponent. Alternative, usedt.strftimeto set a specific string format.strftime()andstrptime()Format Codes-
df.date.dt.strftime('%H:%M')would extract hours and minutes into a string like'14:29' - In the example below, the extracted date is assigned to the same column, but the value can also be assigned as a new column.
-
pandas.DataFrame.pivot_tableis used to aggregate a function if there are multiple values in a column for eachindex,pandas.DataFrame.pivotshould be used if there is only a single value.- This is better than
.groupbybecause the dataframe is correctly shaped to be easily plotted.
- This is better than
- Tested in
python 3.8.11,pandas 1.3.2,matplotlib 3.4.3,seaborn 0.11.2
import pandas as pd
import numpy as np
import seaborn as sns
# create sample data
dates = [f'2016-08-{d}T00:00:00.000000000' for d in range(9, 26, 2)] + ['2016-09-09T00:00:00.000000000']
depths = np.arange(1.25, 5.80, 0.25)
np.random.seed(365)
p1 = np.random.dirichlet(np.ones(10), size=1)[0] # random probabilities for random.choice
p2 = np.random.dirichlet(np.ones(19), size=1)[0] # random probabilities for random.choice
data = {'date': np.random.choice(dates, size=1000, p=p1), 'depth': np.random.choice(depths, size=1000, p=p2), 'capf': np.random.normal(0.3, 0.05, size=1000)}
df = pd.DataFrame(data)
# display(df.head())
date depth capf
0 2016-08-19T00:00:00.000000000 4.75 0.339233
1 2016-08-19T00:00:00.000000000 3.00 0.370395
2 2016-08-21T00:00:00.000000000 5.75 0.332895
3 2016-08-23T00:00:00.000000000 1.75 0.237543
4 2016-08-23T00:00:00.000000000 5.75 0.272067
# make sure the date column is converted to a datetime dtype
df.date = pd.to_datetime(df.date)
# extract only the date component of the date column
df.date = df.date.dt.date
# reshape the data for heatmap; if there's no need to aggregate a function, then use .pivot(...)
dfp = df.pivot_table(index='depth', columns='date', values='capf', aggfunc='mean')
# display(dfp.head())
date 2016-08-09 2016-08-11 2016-08-13 2016-08-15 2016-08-17 2016-08-19 2016-08-21 2016-08-23 2016-08-25 2016-09-09
depth
1.50 0.334661 NaN NaN 0.302670 0.314186 0.325257 0.313645 0.263135 NaN NaN
1.75 0.305488 0.303005 0.410124 0.299095 0.313899 0.280732 0.275758 0.260641 NaN 0.318099
2.00 0.322312 0.274105 NaN 0.319606 0.268984 0.368449 0.311517 0.309923 NaN 0.306162
2.25 0.289959 0.315081 NaN 0.302202 0.306286 0.339809 0.292546 0.314225 0.263875 NaN
2.50 0.314227 0.296968 NaN 0.312705 0.333797 0.299556 0.327187 0.326958 NaN NaN
# plot
sns.heatmap(dfp, cmap='GnBu')
Related videos on Youtube
frankshort
Updated on September 14, 2022Comments
-
frankshort over 1 year
A little info: I'm very new to programming and this is a small part of the my first script. The goal of this particular segment is to display a seaborn heatmap with vertical depth on y-axis, time on x-axis and intensity of a scientific measurement as the heat function.
I'd like to apologize if this has been answered elsewhere, but my searching abilities must have failed me.
sns.set() nametag = 'Well_4_all_depths_capf' Dp = D[D.well == 'well4'] print(Dp.date) heat = Dp.pivot("depth", "date", "capf") ### depth, date and capf are all columns of a pandas dataframe plt.title(nametag) sns.heatmap(heat, linewidths=.25) plt.savefig('%s%s.png' % (pathheatcapf, nametag), dpi = 600)this is the what prints from the ' print(Dp.date) ' so I'm pretty sure the formatting from the dataframe is in the format I want, particularly Year, day, month.
0 2016-08-09 1 2016-08-09 2 2016-08-09 3 2016-08-09 4 2016-08-09 5 2016-08-09 6 2016-08-09 ...But, when I run it the date axis always prints with blank times (00:00 etc) that I don't want. Is there a way to remove these from the date axis?
Is the problem that in a cell above I used this function to scan the file name and make a column with the date??? Is it wrong to use datetime instead of just a date function?
D['date']=pd.to_datetime(['%s-%s-%s' %(f[0:4],f[4:6],f[6:8]) for f in D['filename']])